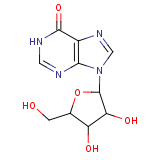
| Compound Information | SONAR Target prediction | | Name: | INOSINE | | Unique Identifier: | SPE03100024 | | MolClass: | Checkout models in ver1.5 and ver1.0 | | Molecular Formula: | | | Molecular Weight: | 256.131 g/mol | | X log p: | 3.243 (online calculus) | | Lipinksi Failures | 0 | | TPSA | 54.26 | | Hydrogen Bond Donor Count: | 0 | | Hydrogen Bond Acceptors Count: | 9 | | Rotatable Bond Count: | 2 | | Canonical Smiles: | OCC1OC(C(O)C1O)n1cnc2C(=O)NC=Nc12 | | Class: | carbohydrate | | Source: | meat extracts, sugar beet, Bacillus subtilis, E. coli, Saccharomyces cerevisiae,
Fusarium spp | | Reference: | Biochem Biophys Res Commun 13:394 (1963); J Pharmacol 14:47 (1983) | | Therapeutics: | cell function activator, cardiotonic |
| Species: |
4932 |
| Condition: |
ARP1 |
| Replicates: |
2 |
| Raw OD Value: r im |
0.7306±0.00494975 |
| Normalized OD Score: sc h |
1.0037±0.000797905 |
| Z-Score: |
0.1866±0.0258638 |
| p-Value: |
0.85197 |
| Z-Factor: |
-7.10594 |
| Fitness Defect: |
0.1602 |
| Bioactivity Statement: |
Nonactive |
| Experimental Conditions | | | Library: | Spectrum | | Plate Number and Position: | 7|D6 | | Drug Concentration: | 50.00 nM | | OD Absorbance: | 600 nm | | Robot Temperature: | 26.50 Celcius | | Date: | 2006-03-23 YYYY-MM-DD | | Plate CH Control (+): | 0.039650000000000005±0.00170 | | Plate DMSO Control (-): | 0.71745±0.00796 | | Plate Z-Factor: | 0.9514 |
|  png png
ps
pdf |
| DBLink | Rows returned: 1 | |
| 5257418 |
9-[3,4-dihydroxy-5-(hydroxymethyl)oxolan-2-yl]purin-6-one; methylmercury; methylmercury |
| internal high similarity DBLink | Rows returned: 2 | |
| active | Cluster 13033 | Additional Members: 5 | Rows returned: 1 | |
|
