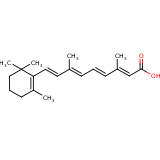
| Compound Information | SONAR Target prediction | | Name: | TRETINON | | Unique Identifier: | SPE01502016 | | MolClass: | Checkout models in ver1.5 and ver1.0 | | Molecular Formula: | | | Molecular Weight: | 272.213 g/mol | | X log p: | 12.087 (online calculus) | | Lipinksi Failures | 1 | | TPSA | 17.07 | | Hydrogen Bond Donor Count: | 0 | | Hydrogen Bond Acceptors Count: | 2 | | Rotatable Bond Count: | 5 | | Canonical Smiles: | CC1CCCC(C)(C)C=1C=CC(C)=CC=CC(C)=CC(O)=O | | Source: | semisynthetic | | Therapeutics: | keratolytic | | Generic_name: | Alitretinoin | | Chemical_iupac_name: | 3,7-dimethyl-9-(2,6,6-trimethyl-1-cyclohexenyl)-nona-2,4,6,8-tetraenoicacid | | Drug_type: | Approved Drug | | Pharmgkb_id: | PA448089 | | Drugbank_id: | APRD00017 | | Melting_point: | 189-190 oC | | H2o_solubility: | 0.6 mg/L | | Logp: | 5.067 | | Cas_registry_number: | 03/08/5300 | | Drug_category: | Antineoplastic Agents; ATC:L01XX22 | | Indication: | For topical treatment of cutaneous lesions in patients with AIDS-related Kaposi-s
sarcoma. | | Pharmacology: | Alitretinoin (9-cis-retinoic acid) is a naturally-occurring endogenous
retinoid indicated for topical treatment of cutaneous lesions in patients with
AIDS-related Kaposi-s sarcoma. Alitretinoin inhibits the growth of Kaposi-s sarcoma
(KS) cells in vitro. | | Mechanism_of_action: | Alitretinoin binds to and activates all known intracellular retinoid receptor
subtypes (RARa, RARb, RARg, RXRa, RXRb and RXRg). Once activated these receptors
function as transcription factors that regulate the expression of genes that control
the process of cellular differentiation and proliferation in both normal and
neoplastic cells. | | Organisms_affected: | Humans and other mammals |
| Species: |
4932 |
| Condition: |
RGP1 |
| Replicates: |
2 |
| Raw OD Value: r im |
0.3669±0.0238295 |
| Normalized OD Score: sc h |
0.8241±0.0451474 |
| Z-Score: |
-4.5859±1.11682 |
| p-Value: |
0.000073518 |
| Z-Factor: |
-0.237806 |
| Fitness Defect: |
9.518 |
| Bioactivity Statement: |
Active |
| Experimental Conditions | | | Library: | SPECMTS3 | | Plate Number and Position: | 8|F4 | | Drug Concentration: | 50.00 nM | | OD Absorbance: | 600 nm | | Robot Temperature: | 26.20 Celcius | | Date: | 2008-06-26 YYYY-MM-DD | | Plate CH Control (+): | 0.039775±0.00066 | | Plate DMSO Control (-): | 0.42974999999999997±0.01289 | | Plate Z-Factor: | 0.8857 |
|  png png
ps
pdf |
| 5460164 |
(2Z,4E,6Z,8E)-3,7-dimethyl-9-(2,2,6-trimethylcyclohexyl)nona-2,4,6,8-tetraenoic acid |
| 6419707 |
(2Z,4E,6Z,8E)-3,7-dimethyl-9-(2,6,6-trimethyl-1-cyclohexenyl)nona-2,4,6,8-tetraenoate |
| 6419708 |
(2Z,4E,6Z,8E)-3,7-dimethyl-9-(2,6,6-trimethyl-1-cyclohexenyl)nona-2,4,6,8-tetraenoic acid |
| 6436389 |
sodium (2Z,4E,6Z,8E)-3,7-dimethyl-9-(2,6,6-trimethyl-1-cyclohexenyl)nona-2,4,6,8-tetraenoate |
| 6439661 |
(2Z,4E,6Z,8E)-3,7-dimethyl-9-(2,6,6-trimethyl-1-cyclohexenyl)nona-2,4,6,8-tetraenoic acid |
| 6439734 |
(2Z,4E,6Z,8E)-3,7-dimethyl-9-(2,2,6-trimethylcyclohexyl)nona-2,4,6,8-tetraenoic acid |
| internal high similarity DBLink | Rows returned: 3 | |
|
