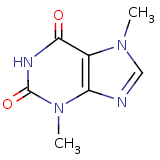
| Compound Information | SONAR Target prediction | | Name: | Theobromine | | Unique Identifier: | LOPAC 00005 | | MolClass: | Checkout models in ver1.5 and ver1.0 | | Molecular Formula: | C7H8N4O2 | | Molecular Weight: | 172.101 g/mol | | X log p: | 1.673 (online calculus) | | Lipinksi Failures | 0 | | TPSA | 52.98 | | Hydrogen Bond Donor Count: | 0 | | Hydrogen Bond Acceptors Count: | 6 | | Rotatable Bond Count: | 0 | | Canonical Smiles: | Cn1cnc2N(C)C(=O)NC(=O)c12 | | Class: | Adenosine | | Action: | Antagonist | | Selectivity: | A1 > A2 |
| Species: |
4932 |
| Condition: |
MT2481-pdr1pdr3 |
| Replicates: |
2 |
| Raw OD Value: r im |
0.6428±0.00480833 |
| Normalized OD Score: sc h |
1.0045±0.00140727 |
| Z-Score: |
0.2578±0.0965724 |
| p-Value: |
0.797038 |
| Z-Factor: |
-4.81577 |
| Fitness Defect: |
0.2269 |
| Bioactivity Statement: |
Nonactive |
| Experimental Conditions | | | Library: | Lopac | | Plate Number and Position: | 15|G8 | | Drug Concentration: | 50.00 nM | | OD Absorbance: | 600 nm | | Robot Temperature: | 27.80 Celcius | | Date: | 2005-04-08 YYYY-MM-DD | | Plate CH Control (+): | 0.04630000000000001±0.00098 | | Plate DMSO Control (-): | 0.6168±0.01715 | | Plate Z-Factor: | 0.8584 |
|  png png
ps
pdf |
| DBLink | Rows returned: 4 | |
| 5429 |
3,7-dimethylpurine-2,6-dione |
| 24700 |
sodium 3,7-dimethylpurine-2,6-dione |
| 205018 |
3,7-dimethylpurine-2,6-dione hydroiodide |
| 3031945 |
lithium 3,7-dimethylpurine-2,6-dione |
| internal high similarity DBLink | Rows returned: 5 | |
| active | Cluster 9605 | Additional Members: 8 | Rows returned: 0 | |
|
